Researchers use new techniques to map vast history of tropical fishes
Jeff Richardson
907-474-5350
Jan. 3, 2024

Caranx ignobilis (giant trevally) aggregate off the southern coast of Mozambique.
An international research team has assembled an unprecedented “tree of life” for a group of more than 150 related tropical fishes, a project that uncovered clues about how distinct species were likely able to evolve over millions of years.
Researchers spent nearly a decade collecting and analyzing DNA data from carangiform fishes, a diverse group that includes species like marlins, billfish, trevallies, remoras and mahi mahis. Along with samples from living species, researchers also used fossils of carangoid species. The effort resulted in the most comprehensive time-calibrated phylogeny to date of those fishes.
The process focused on the emerging science of ultraconserved elements, which are parts of the genome that don’t change as species evolve. Through the analysis of about 900 genes, researchers were able to map out a 56-million-year-old family tree that detailed when various species emerged and how they are related.
“It’s a new tool in the genomics toolbox,” said Jessica Glass, an assistant professor at the University of Alaska Fairbanks College of Fisheries and Ocean Sciences. “No other study has done this for this many species of carangiforms before.”
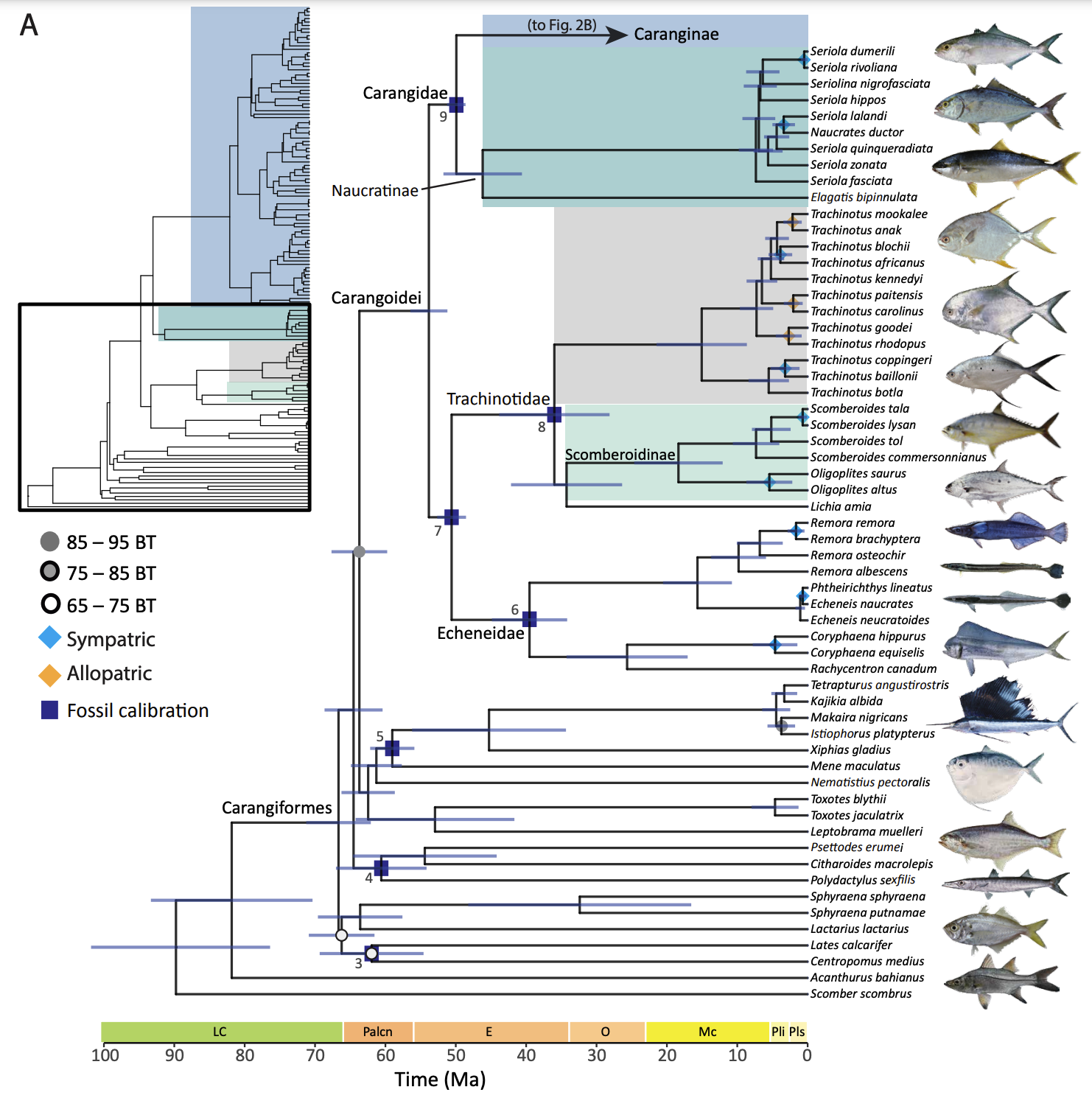
The study also included a deeper look at 41 sister species among carangoid fishes that generally diverged 2 million to 5 million years ago. More than 70% of those species shared the same ranges, conditions that are usually more conducive to intermixing than the evolution of distinct species. But the sister species often existed at different water depths, researchers determined, suggesting that ecological differences allowed them to evolve in overlapping but separate environments.
Glass, who led the study, said having a detailed picture of the evolution of carangoid fishes could be meaningful as the climate warms. As resource managers consider the future of many fish species, a longer look could help them understand what makes them resilient or vulnerable to change.
“If we want to make predictions about how species are going to change over time — even short amounts of time — we can look at how changes in the past affected these species,” Glass said. “That kind of decision-making is better informed when you have an accurate phylogeny.”
The study was published in the November issue of the journal Proceedings of the Royal Society B. Other participating institutions included Yale University, James Cook University, Queensland Museum, Louisiana State University and the South African Institute for Aquatic Biodiversity.
ADDITIONAL CONTACT: Jessica Glass, jrglass@alaska.edu
110-24


